4.5. Example: Li+, Na+, and Cs+ in (C6H6)6
Tip
The sample input and output files can be found in testfiles/rigidmol/2-mben.
4.5.1. Global Optimization
The systems here are benzene-solvated cations: \(\mathrm{Li}^{+}\), \(\mathrm{Na}^{+}\) and \(\mathrm{Cs}^{+}\) in \(\left(\mathrm{C}_6\mathrm{H}_6\right)_{6}\). The parameter files are misc/charmm36/li.xyz, misc/charmm36/na.xyz, misc/charmm36/cs.xyz, and misc/charmm36/c6h6.xyz.
Let’s first consider \(\mathrm{Li}^{+}\).
Step 1: Prepare an input file named li-ben6.inp with the following content:
1li-ben.cluster # cluster file name
220 # population size
320 # maximal generations
43 # scout limit
510.00000000 # amplitude
6li-ben # save optimized configuration
730 # number of LMs to be saved
Step 2: Copy misc/charmm36/li.xyz and misc/charmm36/c6h6.xyz to the current path. Then prepare the cluster file named li-ben.cluster with the following content:
12
2c6h6.xyz 6
3li.xyz 1
4* 10.0000
Step 3: Run the global optimization:
$ rigidmol li-ben.inp > li-ben.out
After a few seconds, You will find the global minimum in li-ben-OPT.xyz and local minima in li-ben-LM.
For \(\mathrm{Na}^{+}\) and \(\mathrm{Cs}^{+}\), the global optimization procedure is similar. Below, the global minima are visualized (li-ben/0.xyz, na-ben/0.xyz, and cs-ben/0.xyz).


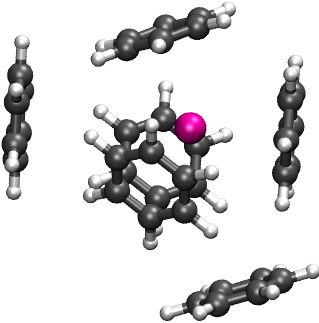
4.5.2. Many-Body Energy Decomposition Analysis
You can also do a many-body energy decomposition analysis (MB-EDA) for these clusters. Its detail can be seen in Theoretical Background and For Molecular Clusters: Many-Body Energy Decomposition Analysis. Here, we just show how to perform MB-EDA for li-ben-OPT.xyz.
You can follow the same procedure given in Example: (H2O)6, giving an input file:
1basis
2 def2-svp
3end
4
5scf
6 charge +1 # Total charge.
7 spin2p1 1
8 type U
9end
10
11grimmedisp
12 type bj
13end
14
15eda
16 type mb_tso
17 mb_level 4
18 frag 0 1 1-12
19 frag 0 1 13-24
20 frag 0 1 25-36
21 frag 0 1 37-48
22 frag 0 1 49-60
23 frag 0 1 61-72
24 frag +1 1 73
25end
26
27mol
28 C -1.75006483 -2.50086495 -1.46649878
29 C -2.81068652 -1.66329526 -1.11472696
30 C -2.62823535 -0.67551816 -0.14457631
31 C -1.38516251 -0.52531075 0.47380254
32 C -0.32454082 -1.36288045 0.12203072
33 C -0.50699198 -2.35065754 -0.84811994
34 H -1.89204211 -3.26951948 -2.22143701
35 H -3.77800267 -1.78018143 -1.59592790
36 H -3.45357435 -0.02375051 0.12916029
37 H -1.24318522 0.24334377 1.22874077
38 H 0.64277533 -1.24599428 0.60323166
39 H 0.31834702 -3.00242519 -1.12185654
40 C 3.51095455 1.53702219 -0.03149057
41 C 3.47459063 2.77605637 -0.67465209
42 C 2.43502071 3.67007163 -0.40967021
43 C 1.43181471 3.32505273 0.49847320
44 C 1.46817863 2.08601855 1.14163471
45 C 2.50774855 1.19200328 0.87665283
46 H 4.31991249 0.84132993 -0.23769046
47 H 4.25525071 3.04453813 -1.38133777
48 H 2.40672359 4.63424502 -0.91015619
49 H 0.62285677 4.02074499 0.70467308
50 H 0.68751856 1.81753679 1.84832039
51 H 2.53604567 0.22782990 1.37713881
52 C 3.35416479 -1.90024170 -1.71678785
53 C 3.01689469 -2.79729695 -2.73252676
54 C 2.66461236 -4.11089136 -2.41542864
55 C 2.64960014 -4.52743052 -1.08259161
56 C 2.98687024 -3.63037527 -0.06685271
57 C 3.33915257 -2.31678086 -0.38395083
58 H 3.62829891 -0.87804723 -1.96354282
59 H 3.02857667 -2.47316066 -3.76969427
60 H 2.40216048 -4.80894860 -3.20584141
61 H 2.37546602 -5.54962499 -0.83583665
62 H 2.97518826 -3.95451156 0.97031480
63 H 3.60160445 -1.61872362 0.40646194
64 C 0.43458154 0.03908166 -3.51702644
65 C 0.56674764 1.10282928 -2.62195220
66 C -0.54649842 1.87721022 -2.28848303
67 C -1.79191058 1.58784355 -2.85008809
68 C -1.92407668 0.52409593 -3.74516233
69 C -0.81083062 -0.25028501 -4.07863150
70 H 1.30087174 -0.56351524 -3.77652080
71 H 1.53588414 1.32800440 -2.18493062
72 H -0.44365132 2.70498169 -1.59196732
73 H -2.65820078 2.19044045 -2.59059373
74 H -2.89321318 0.29892081 -4.18218391
75 H -0.91367771 -1.07805648 -4.77514721
76 C -2.48564491 3.61935571 0.59972959
77 C -3.72351363 3.35244886 0.01097963
78 C -4.64114686 2.52357083 0.65989779
79 C -4.32091137 1.96159965 1.89756592
80 C -3.08304265 2.22850650 2.48631588
81 C -2.16540942 3.05738454 1.83739772
82 H -1.77157395 4.26436040 0.09476356
83 H -3.97270980 3.78975531 -0.95213074
84 H -5.60441333 2.31587319 0.20175299
85 H -5.03498233 1.31659497 2.40253195
86 H -2.83384648 1.79120005 3.44942625
87 H -1.20214295 3.26508217 2.29554252
88 C 1.23249791 -0.30306981 3.95943104
89 C 1.60956722 -1.57772484 3.53134455
90 C 0.63823565 -2.55761031 3.31565089
91 C -0.71016524 -2.26284075 3.52804372
92 C -1.08723456 -0.98818572 3.95613021
93 C -0.11590298 -0.00830025 4.17182387
94 H 1.98835509 0.45944372 4.12727651
95 H 2.65884598 -1.80710431 3.36606782
96 H 0.93165792 -3.54950261 2.98252885
97 H -1.46602242 -3.02535428 3.36019825
98 H -2.13651332 -0.75880625 4.12140694
99 H -0.40932526 0.98359205 4.50494591
100 Li -1.01852524 -0.30118202 -1.75537589
101end
102
103task
104 eda b3lyp
105end
The explaination of this input file can be found in Example: (H2O)6. The only important point is:
Line 6: The total charge is
+1.Line 24: The charge of lithium cation is
+1.
Do the calculation:
$ qbics li-mben-mseda.inp -n 24 > li-mben-mseda.out
Now open li-mben-mseda.out:
1Table 5. Summary (kcal/mol).
2---------------------------------------------------------------------------------------------------------------------------------
3 Interactions delE_el delE_xc delE_pl delE_ct delE_bsse delE_disp delE_tot
4---------------------------------------------------------------------------------------------------------------------------------
5 SUM of 2-body -4.83698400E+01 4.61490292E+01 -4.83508895E+01 -3.37925281E+01 9.22607552E+00 -4.34771067E+01 -1.18615259E+02
6 SUM of 3-body -2.23649606E-08 2.58074315E-01 1.33815835E+01 5.88305181E+00 1.80200373E+00 8.76525397E+00 3.00899673E+01
7 SUM of 4-body 4.13056277E-08 1.59841995E-01 -5.50102044E-01 8.45709613E-01 -2.03912279E+00 -2.67547883E+00 -4.25915201E+00
8 Remain -1.31621640E-08 1.51216649E-02 7.61257854E-02 -2.89732759E-01 4.59645632E-01 3.26936075E-03 2.64429671E-01
9---------------------------------------------------------------------------------------------------------------------------------
10 SUM -4.83698400E+01 4.65820672E+01 -3.54432822E+01 -2.73534994E+01 9.44860210E+00 -3.73840622E+01 -9.25200144E+01
11---------------------------------------------------------------------------------------------------------------------------------
From this table, we can see that the 2-, 3-, and 4-body contribution to the total interaction energy is -118.61, +30.09, and -4.26 kcal/mol, respectively. Since 3-body energy is positive, this term is destablizing the cluster, being anti-cooperative. This is completely different from water clusters in Example: (H2O)6. The anti-cooperativity is mostly contributed from polarization, charge transfer, and dispersion interactions.